|
Research Click on a topic in this outline to jump to the section of interest.
Our laboratory is interested in how genotypic and phenotypic diversity is created during the life cycle of plants.We are studying the regulation of Mutator transposable elements in response to host developmental signals and environmental cues as an entry point.Mutator is regulated by its distribution in maize populations, epigenetically through switches between active and inactive phases, developmentally during tissue ontogeny, and differentially between the somatic and pre-germinal cells.Our "reporter gene" is Bronze2, which encodes the last genetically defined step in anthocyanin synthesis. We are currently analyzing MuDR/Mu biology using transgenic maize expressing individual forms of the MURA transposase combined with MURB transgenes.We also use spontaneous deletion derivatives of MuDR that are missing one of the two genes.Reporter genes include mutable alleles of the anthocyanin pathway and the genetically engineered RescueMu element.RescueMu disrupts a 35S:Lc expression cassette:when the element excises the Lc transcription factor can be expressed, restoring anthocyanin pigmentation.
Function of the MURA and MURB Genes Encoded by MuDR George Rudenko, Akemi Ono, Matthew Fitzgerald Analysis of Mu Transposition Mechanism using RescueMu Gillian Nan, Lily Yu, John Fernandes Mobile Mutator elements are restricted to a few active Mutator lines -- the master element MuDR is not generally found in maize lines, although all maize lines have hMuDR (homologs of MuDR) elements.Mutator transposable elements in maize are organized in a complex family. MuDR (Mutator Don Robertson) has two transcription units encoding MURA and MURB protein products. There are multiple possible protein products from each gene as a result of alternative transcription start sites and alternative splicing decisions. The MURA proteins share similarities with prokaryotic transposases, and an 823 amino acid form of MURA is a sequence-specific DNA binding protein that interacts with a conserved motif within the terminal inverted repeats of mobile Mu elements.In transgenic plants both MURA823 (all 3 introns spliced) and MURA736 (intron 3 retained) can catalyze Mu excision but they fail to catalyze Mu insertion.Alternative splicing of the first mudrA intron followed by frameshift translation can produce an 854 amino acid form of MURA.Both frameshift corrected and frameshift required cDNA constructs catalyze Mu excision, and we are currently documenting the requirements for insertion, i.e. the presence of MURB in addition to MURA854.In vitro all predicted forms of MURB are non-specific DNA binding proteins; this activity could be related to a role of MURBs in selecting new target sites for Mu insertion.Mu elements insert preferentially into genes, but can apparently insert into virtually any gene.Based on particle bombardment assays all forms of MURB localized to the cytoplasm, even in the presence of MURA that was localized to the nucleus; the apparent location of MURBs is inconsistent with a role in target site selection in the chromosomes.To gain more insight into MURB localization and the possible developmental control of compartmentalization, MURB-GFP has been introduced into transgenic maize. hMuDR elements are closely related to MuDR; they are transcribed and produce proteins related to MURA and MURB, but these proteins are incompetent to catalyze transposition. The "non-autonomous" family members share ~215 bp terminal inverted repeats (TIRs) with MuDR, but little internal sequence is shared with MuDR.The Mu1, Mu2, Mu3, and Mu8 slave elements account for the majority of new mutants in active Mutator stocks.
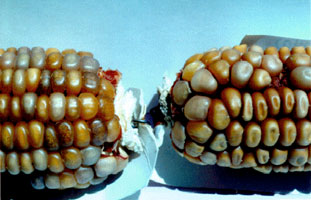
MuDR/Mu elements - developmental control MuDR/Mu elements exhibit two fascinating aspects of developmental control:late timing and a switch in transposition outcome in somatic compared to germinal cells.Mutator elements are active only during terminal cell divisions of tissue development.One hypothesis is that there is direct competition between MURA and transcription factors for binding to the Mu terminal inverted repeats; because assembly of a functional transposasome is slow step and is readily disrupted, Mu excision is delayed until late in development when the transcription factors decrease in concentration.Developmental activation of Mutator is unlikely to be accomplished at the level of transcriptional activation, because MuDR transcripts are ubiquitously expressed. In situ hybridization demonstrates that both sense and antisense MuDR transcripts are found in most cell types at reasonably high levels (probably 100X more message than for Ac transposase, for example).Alternative splicing or translational regulation (perhaps by the abundant antisense transcript encoded by MuDR or by recently discovered 21-26 nucleotide short RNAs) may contribute to the developmental regulation observed. When the Mutator system is finally active late in development, we see element excision and at least a fraction of the excised elements are reinserted by a "cut & paste" mechanism.The "cut only" transposases MURA823 and MURA736 could be responsible for many of the somatic revertant sectors seen in maize tissues.In pre-meiotic cells, during meiosis, and in gametes, Mutator activation results in insertion without element excision.This net replicative transposition in which new insertions are generated without excision at old sites could occur through a "cut & paste" followed by gap repair mechanism or by true replicative transposition as seen in many prokaryotic elements.Using the genetically engineered element RescueMu, we are currently studying the mechanism of germinal insertion. In all somatic tissues examined, Mu element excision occurs late in development, resulting in small sectors. With an anthocyanin reporter gene, these sectors are readily visualized. Shown below are examples of anthers and leaf sheath, both with the bz2::Mu1 reporter allele. Mutator activity is often lost during development, an epigenetic loss because the cryptic elements can be reactivated by radiation. In the inactive phase, the Mu elements acquire a higher level of DNA methylation than Mu elements in active lines. Shown in the photo on the right are 2 ears from the same plant: the ear on the left remained active (1:1 spotted:beige kernels) while the right ear turned off (no spots).
UV-B Activates Silenced MuDR/Mu Elements and Alters the Maize Transcriptome Paula Casati, Virginia Walbot At the whole plant level, we have studied activation of cryptic MuDR/Mu elements during UV-B exposure as an example of how environmental conditions can modulate the pace of genetic change in an organism.UV-B causes extensive DNA damage, an aspect that we have quantified using antibodies to the two most common DNA lesions:cyclobutane pyrimidine dimers and the 6,4 photoproducts.The relationship between transposon activation and DNA damage remains murky, however.To assess more fully the impact of UV-B on maize we have begun using maize microarrays to survey the transcriptome during and after UV-B supplementation, during recovery from UV-B treatment, and after filtering UV-B from sunlight.Our initial experiments demonstrated that anthocyanin is an effective sunscreen in that many responses found after UV-B supplementation to "purple plants" were already present in green plants growing in sunlight.UV-B supplementation decreases the abundance of genes involved in photosynthesis, while genes encoding enzymes important for respiration and glycolysis are increased.Of great interest is the finding that ribosomal protein synthesis increases and that UV-B induces cross-linking between rRNA and proteins.We hypothesize that ribotoxic damage may be as significant as DNA damage and the production of reactive oxygen species.In Mutator plants, mudrA and mudrB transcripts are increased by UV-B; in sunlight, the Mutator plants express higher than normal levels of some repair genes, probably because they experience continuous chromosome breakage from Mu element excision.Current studies are aimed at refining the time course of responses to UV-B using several maize genotypes.We are also pursuing the observation that organs such as immature ears that do not perceive UV-B directly nonetheless show significant transcriptome differences when the plant is treated.Such indirect effects could be responsible for triggering Mutator activity in silenced lines and for the deleterious impact of UV-B on plant growth. Intron Recognition Because plants appear to utilize novel rules for intron recognition, a major effort in the laboratory is discovering the mechanisms of intron recognition using a combination of biochemistry and molecular analysis. Just as we can exploit mutable anthocyanin reporter alleles as sensitive monitors of Mutator activity, we can study post-transcriptional regulation utilizing the Bronze2 gene of this pathway. Using transient assays we are defining the requirements for intron splicing in Bz2 transcripts. One important finding is that specific stresses, given at the whole plant level, can lead to splicing failure in specific transcripts. This suggests that the outcome of intron recognition and processing is highly responsive to environmental conditions. We have designed novel splicing success/splicing failure vector pairs for monitoring the impact of cis mutations in intron processing in transient assays in maize. An important component of this work is a collaboration with mathematician Volker Brendel. Volker's goal is to design methods for precise intron prediction, and together we have developed the concept of local compositional contrast as a means to explain intron recognition in maize.
Sequestration of Anthocyanin: How Does the Pigment Become a Cell-Autonomous Marker? Dean Goodman, Savita Shah Using a combination of biochemistry and genetics we are working on the requirements for anthocyanin sequestration into maize vacuoles. , Early in 1995 we determined that BZ2 is a 26.5 kDa glutathione S-transferase (GST).Recent studies demonstrated that GST activity is not required for maize BZ2 or petunia AN9 to promote the sequestration of anthocyanin into the vacuole.The experimental protocol is to use site-directed mutagenesis to eliminate a residue required for GST enzymatic action, and then bombard bz2 maize aleurone with the altered gene construct.We score anthocyanin sequestration as a purple vacuole in the bombarded region. We propose that a carrier protein is required to move the newly synthesized anthocyanin through the cytoplasm to an MRP pump in the tonoplast; MRPs are a subtype of ABC transporters that use ATP directly to move bulky organic molecules through membranes. An MRP is implicated because anthocyanin sequestration is inhibited by vanadate but not by drugs that discharge the proton gradient across the tonoplast membrane.The carrier protein protects the anthocyanin from oxidation:in the absence of BZ2, maize anthocyanin is oxidized to a brown pigment in the cytoplasm.The carrier protein may also protect the cell from the anthocyanin (and other flavonoids), some of which are mutagenic or bind cellular constituents.A third role of the carrier protein may be to provide a reducing agent for the MRP pump.During in vitro assays of MRP function, reducing agents are required; in vivo, the GST dimer carrying two molecules of anthocyanin would also have four molecules of glutathione.
An important insight from studying anthocyanin sequestration is that the BZ2 function is non-cell autonomous:revertant sectors are surrounded by a paler halo of purple tissue.In contrast, revertant sectors involving the regulatory genes R=B plus C1=Pl are discrete, often just a single cell with a purple vacuole.From these observations we deduced that there is a step after the BZ2 carrier protein function that converts anthocyanin into a cell autonomous marker. Using a bioinformatics approach we identified the MRP genes of maize in EST collections, and then tested each one to determine if expression was regulated by the R=B plus C1=Pl transcription factors regulating the anthocyanin biosynthetic pathway.MRP29 is strongly up-regulated along with the anthocyanin pathway in various maize tissues. MRP29-GFP fusion proteins are localized to the maize tonoplast.Therefore, the gene is expressed in the correct tissues and the protein is localized to the appropriate compartment to facilitate anthocyanin sequestration.In genetic analysis, however, Mu disruptions of MRP29 did not "knockout" anthocyanin sequestration.Antisense-MRP29 constructs did create a "beige" tassel phenotype suggestive of anthocyanin oxidation in the cytoplasm; by HPLC the antisense plants contained the appropriate anthocyanins.Our current working hypothesis is that there is more than one MRP competent to pump anthocyanin into maize vacuoles.
Maize Gene Discovery Project: Tagging Maize Alleles with RescueMu and DNA Sequencing of ESTs and Mutant Alleles Gillian Nan, Bret Schneider, Darren Morrow, Brian Nakao, John Fernandes, Diane Chermak As part of the Maize Gene Discovery Project we are constructing RescueMu tagging grids and we are processing the row and column DNA preps from project grids to generate templates for DNA sequencing. As a second method for maize gene discovery, we are sequencing Expressed Sequence Tags (ESTs). The genomic sequences adjacent to RescueMu insertion sites are assembled with maize ESTs to permit identification of introns and the 5' and 3' non-coding regions. The genomics projects are conducted at the Stanford Genome Technology Center, and the sequence data are annotated by ZmDB. Analysis of the RescueMu data permits us to evaluate many aspects of Mutator biology previously reported from a few examples; we can also discover new aspects of Mu element behavior. For example, we can estimate deletion frequency within and adjacent to these elements, and we can assess the targets of Mu insertion on a genomics scale. Results of study on organ-specificity of gene expression patterns using ESTs and microarray analysis from a manuscript for Plant Physiology EST Project: We have greatly exceeded our goal to sequence 50,000 ESTs drawn from libraries prepared by the project participants and donated by members of the maize genetics community. As of April 2002, we have deposited 110,000 ESTs in GenBank. Transposon Tagging All Maize Genes: Grids will be organized of 2304 plants (48 rows of 48 individuals each) that have been tagged with RescueMu, a derivativeof the Mu1 transposon found in Mutator lines of maize. Pools of leaf punches will be collected from each row and from each column to generate 96 libraries of RescueMu insertions. Genomic DNA Sequencing: Plasmid rescue of RescueMu and the adjacent genomic DNA from tagging populations will create a permanent collection of the insertion mutations. Virtually all Mu element insertions are into or near genes so sequencing 1.2 kb flanking >150,000 insertions should provide the genomic sequences of the expected 50,000 maize genes with 95% probability. We will sequence from the row libraries only. As of April, 2002, we have sequenced grids G, H, and I, and we will be sequencing a new grid approximately every three months until the end of the project." Gene Annotation Tools Tailored To Maize: A major bioinformatics goal of our project is to use new, original research on intron definition and new statistical tools to improve prediction of gene structure from genomic sequences. EST sequences will also be important in this effort.
Phenotypic Analysis of Transposon-Tagged Mutants Microarray ESTs and Genomic Sequences To aid the phenotypic characterization of maize genes, all sequenced items will be microarrayed onto glass slides suitable for gene expression studies. The project team will conduct survey experiments to define the patterns of gene expression in the major organs of maize, to compare Mutator and non-Mutator lines of maize, and to compare inbred lines used in the transposon tagging populations. Phenotypic Analysis of Mutator Lines The RescueMu tagging individuals will be self-pollinated, and the progeny will be evaluated for novel phenotypes at the kernel, seedling, young adult, and floral stages. Phenotypic records and photographs will be provided for each tagging individual, organized into a row composed of 48 individuals. The DNA sequences of the RescueMu elements from a row of individuals will be reported with that row. As of April, 2002, we are distributing microarrays of PCR-amplified cDNAs drawn from four EST sequencing projects (~3,000 different genes on each array type) and the first Unigene arrays (~6,000 genes on Unigene1.1).
New Tools for the Maize Genetics Community Distribute
the RescueMu Plasmid Collections in Indexed Sets Distribute
Transposon Tagged Lines Develop
a New DNA Hybridization Gene Mapping Method |